The GRCh37 assembly in Ensembl
The human assembly GRCh37 (also known as hg19) in Ensembl is available as a stable archive, so that you can continue to analyse your human data on its previous sequence. To browse genes, variants and genomic regions all assigned with the previous genomic coordinates, visit our GRCh37 dedicated site.

Ensembl tools are also available for analyses using the old GRCh37 assembly. For example, you can annotate your variants with VEP for human GRCh37, export Ensembl annotation on GRCh37 with BioMart, and run BLAST/BLAT similarity searches against the GRCh37 assembly.
The data and annotation on GRCh37 can also be downloaded as MySQL databases and file dumps from our FTP site. You can also access Ensembl data on GRCh37 via the REST API.
This stable archive will be updated twice a year to incorporate new data such as SNPs and phenotype from dbSNPs and NHGRI-EBI GWAS, in addition to a few other datasets from different databases. Check our Ensembl blog for the news on the Ensembl GRCh37 updates.
The gene annotation on the GRCh37 archive is based on Ensembl data from release 75 and will not be updated. For the latest annotation of genes, variation, comparative genomics and regulatory data, please use the main Ensembl site. This site and its underlying databases are on GRCh38, the latest assembly of the human genome.
GRCh38 is an improved representation of the human genome compared to GRCh37, where many gaps were closed, sequencing errors corrected and centromere sequences modelled. For the state-of-the-art of the human genome and its annotation, go to GRCh38.
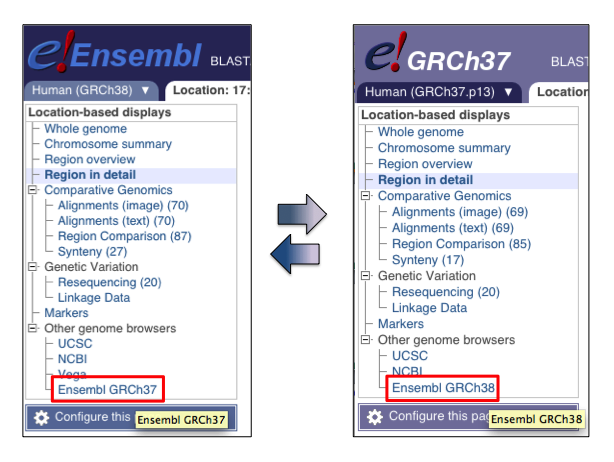
Converting GRCh37 into GRCh38
If your data is in GRCh37, you can swiftly move to GRCh38 genomic coordinates:
- navigate from the GRCh37 site to the corresponding GRCh38 location on our main website (see figure, below)
- convert GRCh37 to GRCh38 coordinates using our assembly map endpoint on the REST API service
- use our Assembly Converter tool to process multiple regions at a time.
We anticipate that the majority of our users will be migrating to GRCh38, as it contains the most updated and reliable annotation of the human genome.
