Sequence
This page shows the genome sequence for a gene of interest. Flanking sequence and introns are displayed. Exons belonging to the gene of interest are shown in red letters with peach highlighting. Other exons within this region that do not belong to the gene of interest are shown in black letters with peach highlights. Note that coding and non-coding transcripts contribute to this view.
Change the display and highlight features in the gene structure using the Configure this page options as follows:
- Flanking sequence Select the extent of the flanking regions upstream/downstream of the gene (default is 600 bps).
- Number of base pairs per row Change the number of base pairs displayed in each row.
- Additional exons to display Exons from Ensembl (core exons), Vega (manually annotated), ab initio or EST gene exons (gene models based on EST evidence, EST genes) can be highlighted.
- Orientation of additional exons Exons on the forward, reverse or both strands can be highlighted.
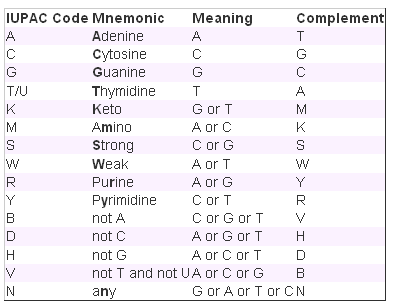
- Show variations All variations (with links) or no variations can be shown. These will be indicated as IUPAC codes (find a table at the bottom of this page) within the sequence. Click a variant within the sequence to see the strand. (For the sequences on the reverse strand, the ambiguity code will also be relative to the reverse strand).
- Filter variations by population Choose a specific population to view.
- Minor allele frequency for population filter Demand a cut-off for variation frequency within a population
- Line numbering Relative to the sequence (starting at the number 1 in the sequence displayed), relative to the coordinate system (genomic coordinates) or no numbering can be selected.
To BLAST sequence, select your sequence of interest with your mouse then click on the pop-up button.
Clip: Genome Variation
IUPAC Ambiguity Codes